Mapping the Genetic Code of Wild Oat Species
Oats have been domesticated more than 3,000 years ago while growing as a weed in wheat fields in the Anatolian region of Turkey. Their genomes are rich in genetic diversity, as shown by the large genome size (12.6 Gb, 6x) and the mosaic-like chromosomes. Little is known about the evolutionary events that lead to structural variation in oat chromosomes. However, the diploid wild oat species have a similar number of genes and are the gene bank of stress resistance, adaptive characters of oats. To this end, Dr. Liu Qing’s team of the Rare and Endangered Plant Conservation Research Group assembled the genome sequence of wild oat species and conducted the comparative genomics research. The research results deepened people’s understanding of the oat genome origin.
Recently, the article was published on Scientific Data entitled “Chromosome-level genome assembly of the diploid oat species Avena longiglumis”. The combination of Illumina, Nanopore and Hi-C data allowed researchers to assemble a high-quality chromosome-level genome of Avena longiglumis (Genome size 3,960.97 Mb). A total of 40,845 protein-coding genes were annotated (Figure 1). The transcriptome data of salt-treated oats were obtained, which provided new implications for the development of salt-tolerance genes in oats. Dr. Qing Liu (First author) and Prof. John Seymour Heslop Harrison are the co-corresponding authors. Dr. Tieyao Tu, Dr. Ziwei Wang (Current affiliation is School of Biology and Agriculture, Shaoguan University), and master student Gui Xiong of South China Botanical Garden participated in this work.
Comparative genomics studies have found that the collinear regions were highly conserved in wild oat species A. longiglumis and A. eriantha (2n = 2x = 14). The terminal gene-rich chromosomal segments (representing about 50 Mb) showed translocations between chromosomes during the speciation of diploid wild species (Figure 2), and the newly-formed intergenomic translocations of similar extent were found in the hexaploid A. sativa. The post-rho polyploidization events were verified to occur in Avena. The study provided insight into evolutionary mechanisms and speciation in the Pooideae grasses, which is valuable for the exploitation of oat wild germplasm resources through breeding programs in Poaceae (BMC Plant Biology, 2023, 23:627).
The above researches were funded by the National Natural Science Foundation of China, the Guangdong Basic and Applied Basic Research Fund, and the South China Botanical Garden Overseas Distinguished Scholar Project.
Contact:Associate Prof. LIU Qing,South China Botanical Garden, Chinese Academy of Sciences,E-mail: liuqing@scib.ac.cn
Article links:
Liu et al. Scientific Data, 2024, https://doi.org/10.1038/s41597-024-03248-6
Liu et al. BMC Plant Biology, 2023, https://doi.org/10.1186/s12870-023-04644-7

Figure 1. Genomic characterization of Avena longiglumis
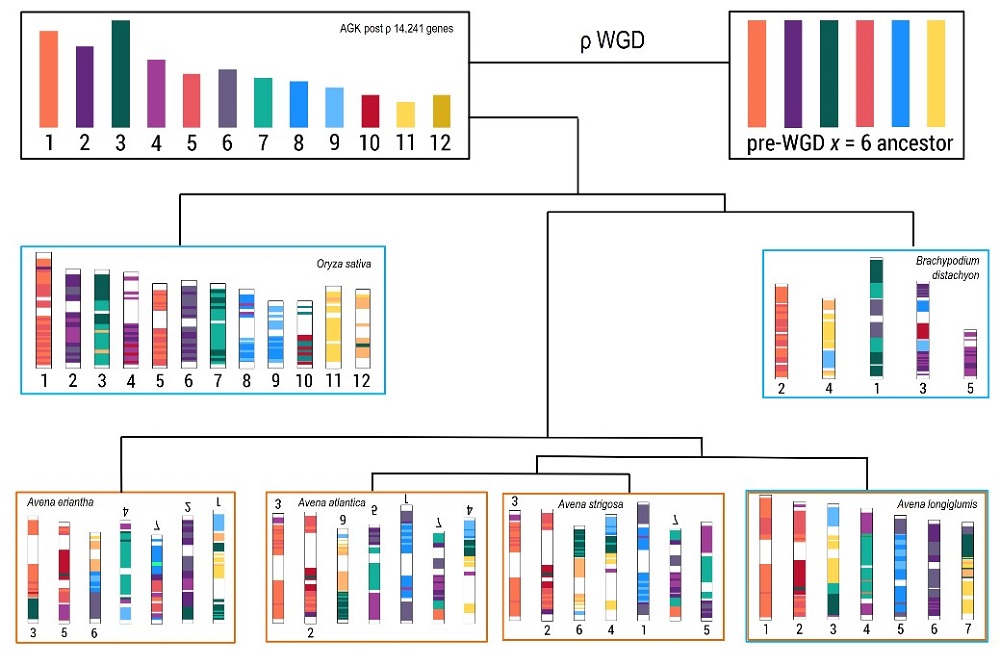
Figure 2. Reconstruction of ancestral grass karyotype for the Pooideae grasses
File Download: