Uncover the molecular mechanism underlying heterostyly in Rubiaceae: Auxin gene identified as a key “switch”
Scientists have discovered that the diversity of flowers in flowering plants may largely stem from their interactions with pollinators. To attract these pollinators, flowering plants have evolved a wide array of floral structures and morphologies during evolution. Interestingly, even distantly related plants can develop similar floral traits in response to similar pollination environments—an example of convergent evolution. However, whether the genetic basis behind such convergent floral traits is also shared remains one of the key questions in evolutionary biology. Heterostyly is a striking example of floral polymorphism: within the same species, two or three floral morphs coexist, with reciprocal positioning of stigmas and anthers (long styles paired with short anthers, and vice versa). This reciprocal arrangement enhances precise pollen transfer, promotes outcrossing, maintains population diversity, and reduces both pollen interference and inbreeding. Heterostyly has been identified in at least 28 angiosperm families, with the Rubiaceae family showing the highest number of genera and species exhibiting this sexual system. However, the genetic and molecular mechanisms underlying heterostyly remain poorly understood.
The research team led by Dr. Shixiao Luo at the South China Botanical Garden of the Chinese Academy of Sciences focused on the genus Mussaenda (Rubiaceae) to explore this question. By integrating de novo genome assembly, comparative genomics, population genomics, transcriptomics, and phylogenetics, the team identified the structure and evolutionary origin of the S-locus supergene that controls heterostylous development in Mussaenda. The key findings include: The supergene controlling heterostyly in Mussaenda consists of three tightly linked genes (MuIAA, MuGA3ox, and MuAPs), and is present only in short-styled individuals, exhibiting a hemizygous structure; Due to recombination suppression caused by the hemizygous structure, the S-locus region is significantly enriched with repetitive sequences compared to flanking regions; The supergene likely originated through stepwise gene duplication and insertion: MuIAA duplicated before the diversification of Rubiaceae, while MuGA3ox was duplicated after species divergence within the family; The auxin-responsive gene MuIAA is highly expressed in the style and floral tube of short-styled flowers, making it a strong candidate gene for regulating style length.
This study not only demonstrates a convergent molecular mechanism underlying heterostyly evolution (i.e., the common hemizygous structure of the S-locus), but also, for the first time, highlights the central role of auxin-related genes in regulating style length. Future research questions include: Why are certain plant lineages more prone to evolving heterostyly? How do hormonal pathways contribute to the maintenance of such complex floral traits? These findings open new windows into understanding the origins of plant diversity.
The research article, titled "Genomic evidence unveils the genetic architecture and evolution of the S-locus controlling heterostyly in Rubiaceae," was recently published in New Phytologist (IF5-year = 10.3). Dr. YUAN Shuai, Associate Researcher at South China Botanical Garden, is the first author, with Prof. LUO Shixiao and Prof. ZHANG Dianxiang as co-corresponding authors. The study was supported by the National Natural Science Foundation of China and the Young Talents Program of the South China Botanical Garden.Article link:https://nph.onlinelibrary.wiley.com/doi/10.1111/nph.70311

Figure 1. Images of the three distylous Mussaenda species investigated in this study. (a, b) Mussaenda lancipetala; (c, d) Mussaenda multinervis; (e, f) Mussaenda macrophylla. In the floral dissection images, the long-styled flower is shown on the left and the short-styled flower on the right. Bars, 1 cm.
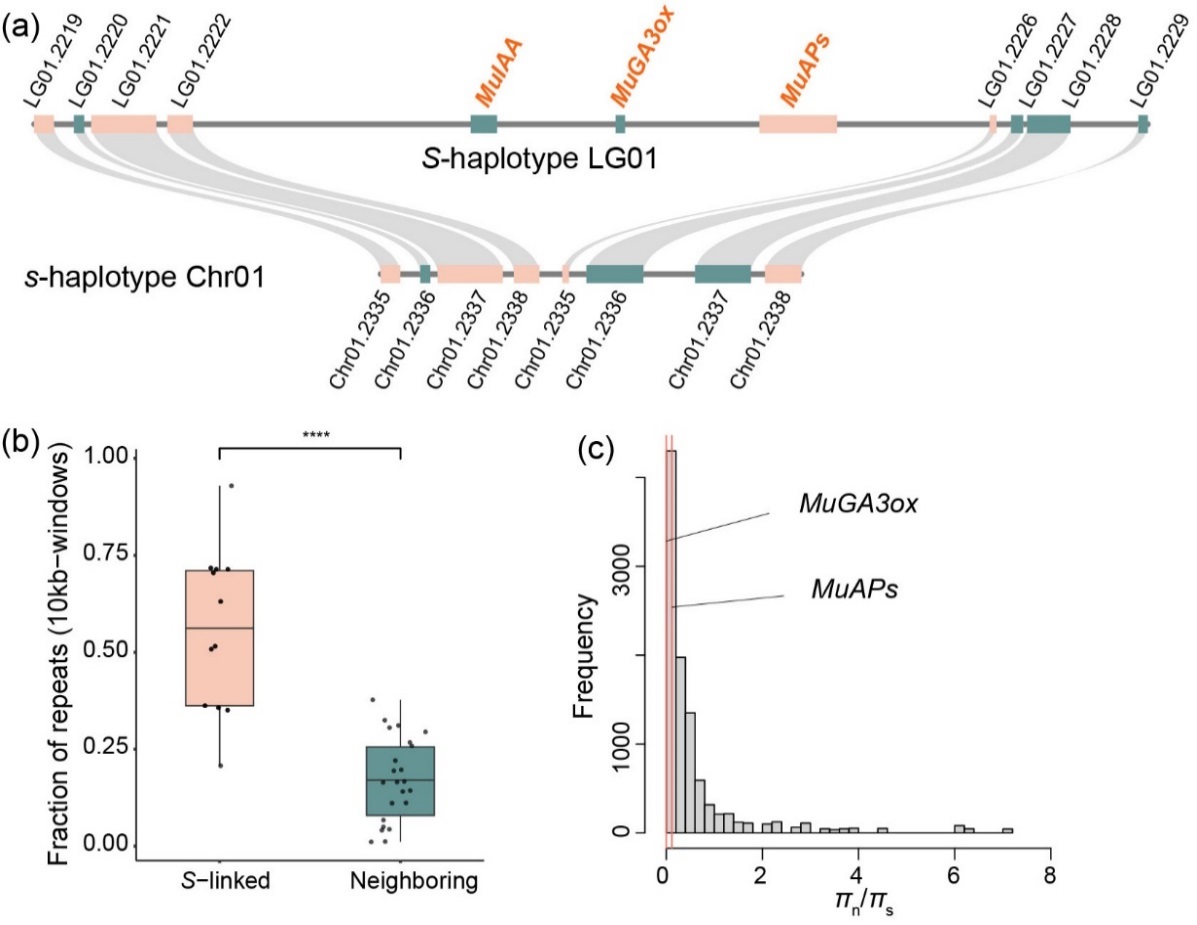
Figure 2. Hemizygous structure and molecular evolution patterns of the S-locus genes in Mussaenda lancipetala. (a) Microsynteny analysis between the genomes of the S-haplotype and s-haplotype indicating the hemizygosity of S-locus genes, which are only present in the S-morph. (b) Comparison of repetitive sequence enrichment in the 120-kb S-locus region (46.55–46.67 Mb) and the adjacent upstream and downstream 12 windows (10 kb each). (c) πn/πs of the S-locus genes in the S-morph genome (n = 33) compared with 10 000 bootstraps of resampling from 100 genes upstream and 100 genes downstream. The πn/πs values of the S-locus genes fall within the 95% confidence interval of the resampled distribution.
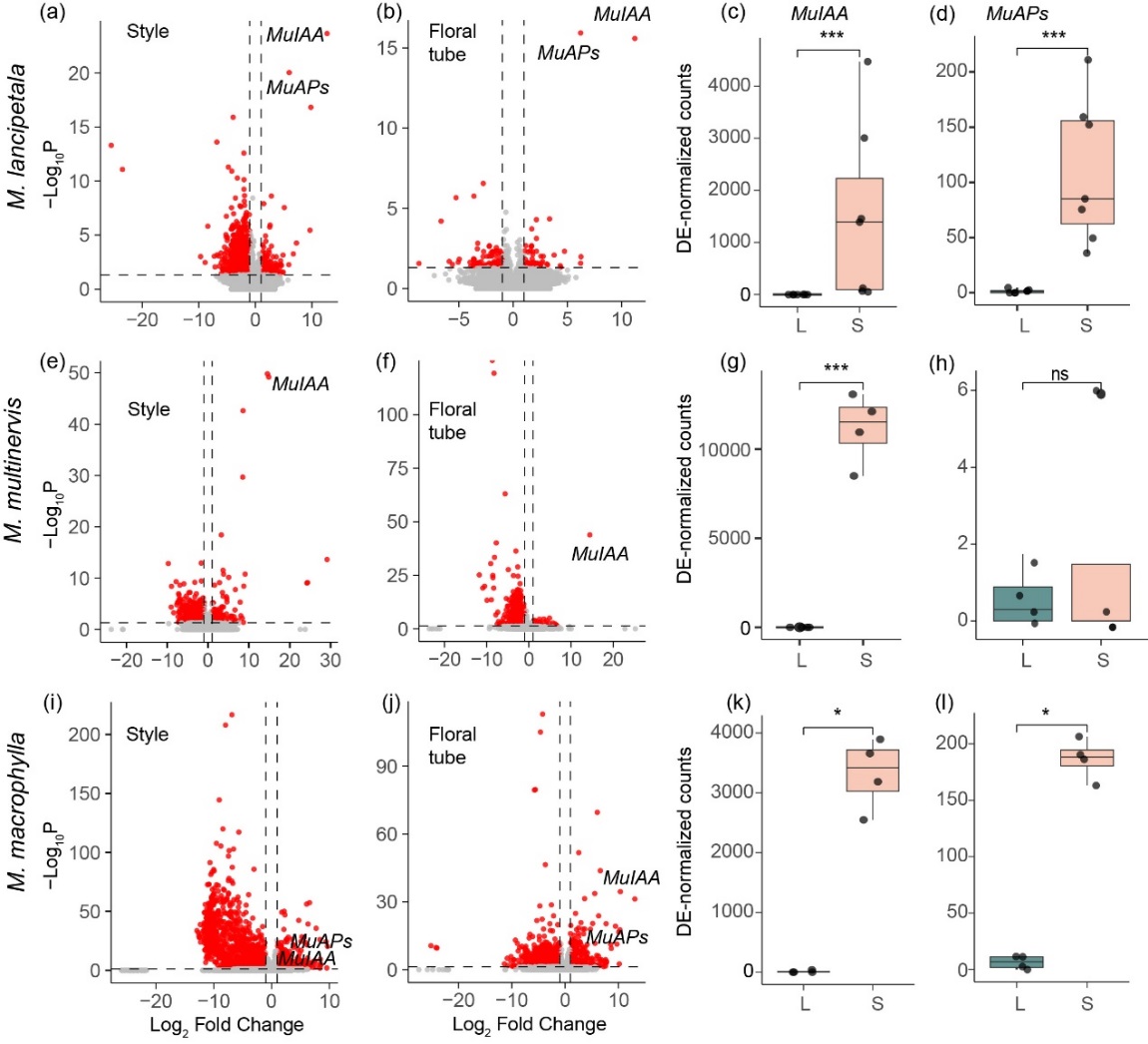
Figure 3. Differential gene expression of flowers of the long (L)- and short (S)-styled morphs of Mussaenda lancipetala, M. multinervis, and M. macrophylla showing S-locus genes that are specifically expressed in the S-morph. The volcano plots display log2-fold change in expression vs significance for (a) pistils and (b) floral tube (filaments) of M. lancipetala, (e) pistils and (f) floral tube (filaments) of M. multinervis, and (i) pistils and (j) floral tube (filaments) of M. macrophylla. Genes differentially expressed between L- and S-morphs are highlighted in red (P adjusted < 0.05). The horizontal dashed line represents fold-change threshold of |1|. In (c, d, g, h, k, l), we compare the normalized counts of MuIAA and MuAPs between pistils of the L- and S-morph.
File Download: